

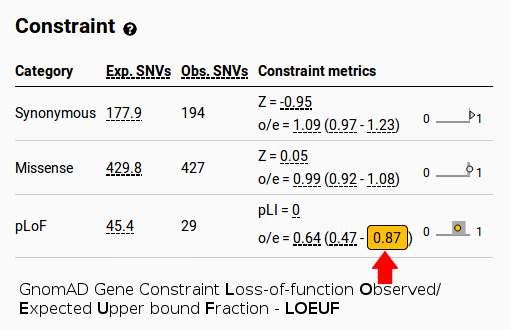
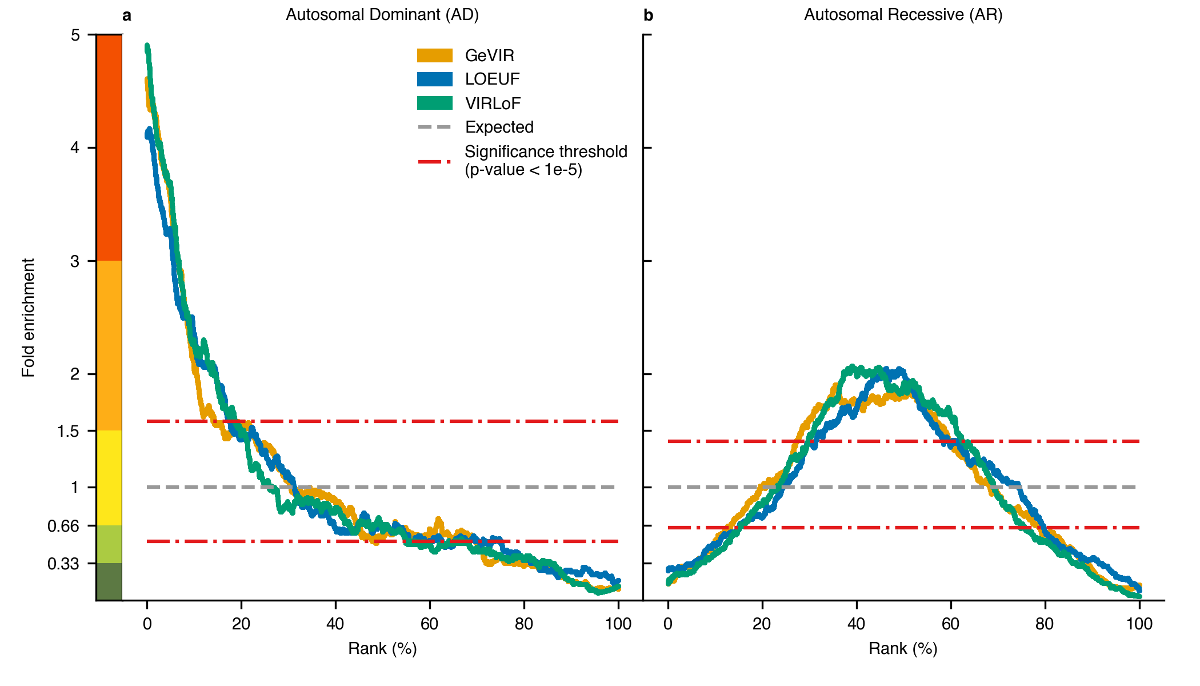
|
|
|
|
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|---|---|---|
| TRPC1 | ENST00000476941 | 10.08 | 23.94 | 14.78 | 2.04 | 1.38 | 1.95 | 0.53 | 0.95 | 0.62 |
| BCL10 | ENST00000370580 | 10.08 | 37.93 | 22.13 | 2.02 | 0.78 | 1.16 | 0.54 | 1.70 | 0.97 |
| MFSD2A | ENST00000372809 | 10.09 | 25.79 | 15.77 | 2.02 | 1.23 | 1.77 | 0.54 | 1.05 | 0.66 |
| BLOC1S2 | ENST00000370372 | 10.09 | 67.80 | 37.24 | 2.02 | 0.52 | 0.82 | 0.54 | 1.18 | 1.99 |
| MSL1 | ENST00000579565 | 10.10 | 2.99 | 3.88 | 2.02 | 3.47 | 3.75 | 0.54 | 0.28 | 0.23 |
| FNTB | ENST00000246166 | 10.10 | 22.17 | 13.78 | 2.02 | 1.52 | 2.07 | 0.54 | 0.81 | 0.57 |
| ENC1 | ENST00000302351 | 10.11 | 23.22 | 14.38 | 2.02 | 1.43 | 2.01 | 0.54 | 0.92 | 0.59 |
| ZNF287 | ENST00000395824 | 10.11 | 18.50 | 11.92 | 2.02 | 1.49 | 2.28 | 0.54 | 0.76 | 0.46 |
| ARHGEF12 | ENST00000397843 | 10.12 | 4.66 | 4.73 | 2.02 | 3.26 | 3.68 | 0.54 | 0.31 | 0.25 |
| RP11-211G3.3 | ENST00000449623 | 10.12 | 83.84 | 45.67 | 2.02 | 0.28 | 0.68 | 0.54 | 0.47 | 1.99 |
| VWC2L | ENST00000312504 | 10.13 | 45.35 | 25.88 | 2.02 | 0.68 | 1.01 | 0.54 | 1.99 | 1.23 |
| AZIN1 | ENST00000337198 | 10.13 | 8.63 | 6.74 | 2.02 | 2.49 | 3.00 | 0.54 | 0.47 | 0.32 |
| PANK3 | ENST00000239231 | 10.14 | 48.73 | 27.56 | 2.02 | 0.63 | 0.96 | 0.53 | 1.99 | 1.27 |
| SF3B2 | ENST00000322535 | 10.14 | 7.03 | 5.84 | 2.02 | 2.57 | 3.30 | 0.53 | 0.37 | 0.28 |
| NUPL1 | ENST00000381736 | 10.15 | 15.18 | 10.23 | 2.02 | 1.90 | 2.21 | 0.53 | 0.67 | 0.42 |
| ELF3 | ENST00000359651 | 10.15 | 13.64 | 9.34 | 2.02 | 2.05 | 2.32 | 0.53 | 0.59 | 0.38 |
| PPP2R2B | ENST00000336640 | 10.16 | 12.32 | 8.57 | 2.02 | 2.07 | 2.52 | 0.53 | 0.59 | 0.36 |
| ATP6V1F | ENST00000492758 | 10.16 | 83.63 | 45.61 | 2.01 | 0.27 | 0.68 | 0.54 | 0.47 | 1.98 |
| EHBP1 | ENST00000263991 | 10.17 | 14.90 | 10.11 | 2.01 | 1.94 | 2.24 | 0.54 | 0.62 | 0.42 |
| CSNK1G1 | ENST00000303052 | 10.18 | 6.83 | 5.77 | 2.02 | 2.62 | 3.31 | 0.54 | 0.37 | 0.28 |
| SLC41A1 | ENST00000367137 | 10.18 | 20.83 | 13.10 | 2.02 | 1.43 | 2.10 | 0.54 | 0.83 | 0.52 |
| FAM46A | ENST00000320172 | 10.19 | 16.91 | 11.20 | 2.02 | 1.64 | 2.15 | 0.54 | 0.71 | 0.45 |
| PLP1 | ENST00000418604 | 10.19 | 16.19 | 10.79 | 2.02 | 1.81 | 2.19 | 0.54 | 0.70 | 0.43 |
| SNX27 | ENST00000368843 | 10.20 | 5.16 | 4.95 | 2.02 | 3.20 | 3.69 | 0.54 | 0.32 | 0.25 |
| GLTSCR1L | ENST00000314073 | 10.20 | 0.39 | 2.75 | 2.02 | 4.14 | 4.03 | 0.54 | 0.28 | 0.23 |
I agree Our site saves small pieces of text information (cookies) on your device in order to deliver better content and for statistical purposes. You can disable the usage of cookies by changing the settings of your browser. By browsing our website without changing the browser settings you grant us permission to store that information on your device.